Making friends with our microbiome
Dear Friends
How are recent analytical advances within the field of microbiology bringing our relationship with Mother Nature back centerstage into the practice of medicine?
Part and partial to the practice of medicine is accurately understanding our relationship with the microbial world—that unseen world that became visible through the world of Louis Pasteur. His work in the late 19th Century brought germ theory into the field of medicine. His focus was on bad bugs and that they must be killed—pasteurization and hygeine became good medical standards of practice.
But, we have come along way since then. NIH, through the Human Genome Project, in collaboration with scientists around the world, developed advanced analytical tools in order to map the human genome, and these tools became a platform through which they could take on the analysis of the microbial world that lives in us and on us—the human microbiome. In 2007 NIH began the Human Microbiome Project with a goal of determining the core microbiome of a healthy human being.
In this week’s Forward Thinking we will share the insights expressed by Gevers D, Knight R, Petrosino J et al. in their paper, The Human Microbiome Project: A Community Resource for the Healthy Human Microbiome. (2012) PLos Biol 10(8): e1001377. In particular, we will look at two new analytical tools used in the leading edge of human microbiology—High Throughput 16S Sequencing and Shotgun Metagenomic Analysis.
The bottomline of what we are discovering in that good microorganisms in our gut and on our body are instrumental to our health and wellbeing. They are partners in our physiological functioning, they are soldiers in our immune systems, they are crucial to our very survival. To find out more about the tools to analyze who and what these microbial friends of ours are read more.
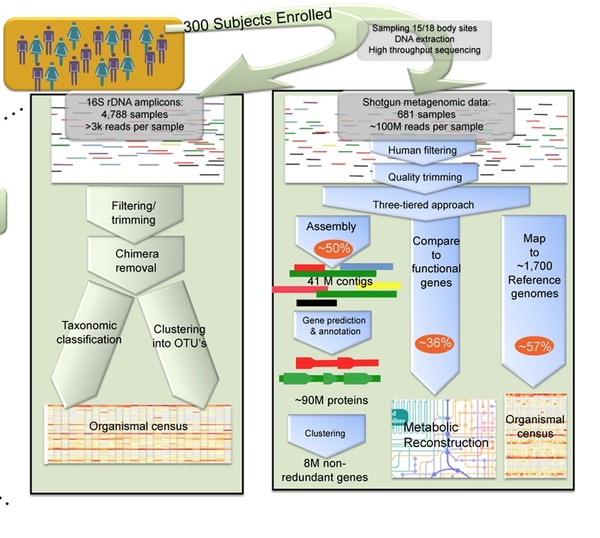
In the above diagram, taken from Govers D et al, we can see the two methods of analysis—on the left 16S rDNA Amplification and on the right Shotgun Metagenomic reads. The challenge had been for microbiologist to see the microbes that live in the gastrointestinal tract for example—95% are unculturable outside of the body. With these new technologies scientists are able to identify and study the bugs in sito.
The procedures are as follows: First, you take a sample from one of the body sites—for example, the stool which is used for determining the bugs of the GI tract. Then, researchers pop the cells open and get the total DNA content. In some samples there will actually be human DNA in there mixed bacterial DNA because of the nature of the body site and how the sample was collected. Finally, through the use of PCR primers, scientists can amplify (make multiple copies) of the 16S rRNA fragments so that we can sequenced for their bases. This provides us with the bacterial fingerprint for each organism, for 16S rRNA sequencing takes us down to the species level of bacterial identification. Much of the work now is to achieve a complete genomic mapping of the bugs in the gut. The HMP consortium has already completed the sequencing of 1000 organisms, resident to the gastrointestinal tract.
Next, in regards to Shotgun Metagenomic Analysis, scientists take the original sample and just shear it into little pieces. Then, they sequences the DNA fragments into genes. The low hanging fruit that comes out of the shotgun approach is that you can get the entire gene content of the sample which allows you to map those genes back to the known metabolic pathways that exist in a bacterial cell. This gives you an idea of the metabolic potential of a given sample. This allows you to do a basic metabloic reconstruction of the sample. How well does it metabolize carbohydrates, or polysaccharides, amino acids, etc. You compare from sample to sample to get an idea of the different metabolic pathway potentials of a give sample.
How does all of the above relate us to Mother Nature? Mother Nature is the biosphere. The biosphere is the tree of life. Within the tree of life we are the branches of the evolutionary process and the microbial world our foundation. You could say we are its offspring. The Human Microbiome Project is making it visibly clear just how marvelously connected we are to the microbial world. We all Mother Nature, the microbial world and ourselves are one.
A very big hug to you all and Happy Holidays from Dohrea and me!
Sincerely yours, Seann Bardell
Clinical Note:
The BioImmersion Synbiotic Formulas are the Original Synbiotic formula, the Beta-Glucan Synbiotic Formula, the Triple Berry Probiotic Formula, the High ORAC Synbiotic Formula, the Supernatant Synbiotic Formula, the Cranberry Pomegranate Synbiotic Formula and the No. 7 Systemic Booster. They represent the following strains: L. bulgaricus ATCC pending, DUP 14073, L. helveticus ATCC 7994, L. casei ATCC 393, B. infantis ATCC 15697, B. longum ATCC 15707, L. acidophillus ATCC 4356, S. thermophillus ATCC 19258, L. plantarum ATCC 8014 and L. rhamnosus ATCC 7469.
We will be driving into the research on the different strains, relative to human health.
![]()

This is the season for giving and in this article collected by The Global Oneness Project called Radical Generosity, Paul Van Slambrouck explores the diversity of gift economy projects around the globe. It is very thought provoking and inspiring. Everyone can do it, all can participate. Check it out.